
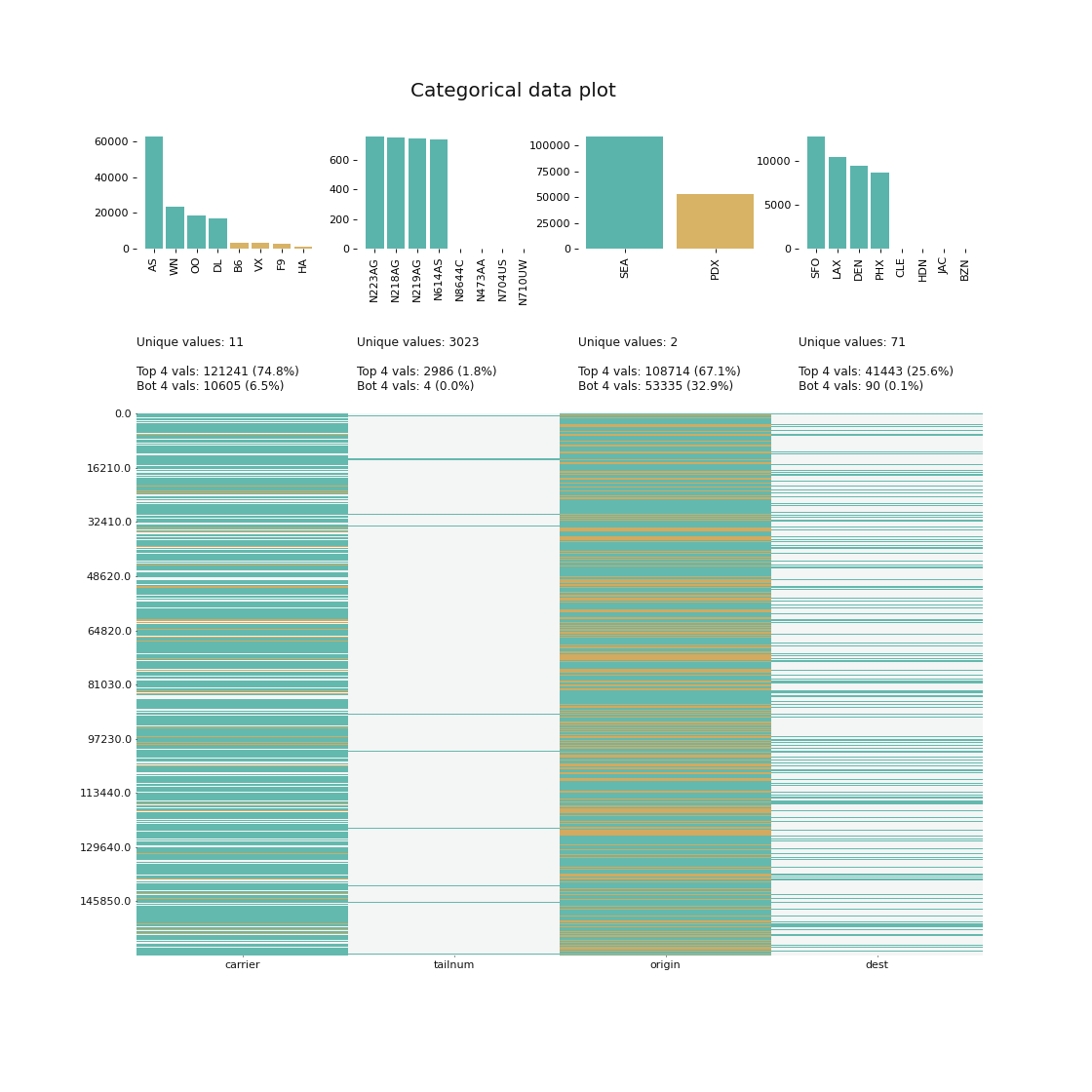
If anyone can fix the CodecZlib issue, that would be great. After this, Julia is about 1.4 slower than C on gzipped data - whereas we ought to be very near C speed here, something like a factor of 1.1. I created this issue - Jaakko Ruohio on Slack discovered about half the problem was that zlib was not compiled with -O3, but there are still more gains to be had, I just don’t know where to find them. Profiling confirms that almost all time is spent in the ccall line. That’s very strange, considering it just calls zlib directly. Not bad! We probably shouldn’t actually remove the boundschecks, because IMO the extra safety is worth a little extra time, especially considering that FASTQ files are essentially always gzipped.įor the gzipped data, it appears that CodecZlib is very slow, something like 4x slower than zlib. With this optimization, the running speed of Julia is about 1.25-1.3 times as long as his C code. Since it reads the input byte by byte, this has a rather large effect, taking another 30% time off the non-zipped input.

There’s some type instability, but union splitting takes care of that, and it’s no performance concern.įor the FASTX code, I noticed the FASTQ parser does not use inbounds. Looking at his Julia code, it looks great. It might be a matter of hardware, or the fact that I ran FASTX v 1.1, not 1.0. I don’t see the same relative numbers b/w C, Python and Julia that he does - for me, Julia does about 30% better on non-zipped data. It’s much more interesting to look at the FASTQ benchmark using FASTX.jl from BioJulia.
#Klib library python code#
I don’t think it’s useful to pick that code apart, it’s just one random guy’s Julia code. The library Klib.jl is a bit obscure, seemingly just used by one guy, and not very well maintained. recreated the C, Julia and Python part of the first benchmark. The outcome is a (legit) cyclic dependency error.

Run the cinteropGles31 task again Again the task should not run, but gradle notices that the klib files has changed, and since it's somehow an input of this task, it runs again. 0 Note changes in depends and also includeHeaders and exportForwardDeclarations. 71 abi_version= 22 exportForwardDeclarations= At this point klib/manifest has become: unique_name=effects-backend-cinterop-gles31ĭepends=stdlib posix glesCommon effects-backend-cinterop-gles31Ĭompiler_version= 1.3. I really don't know how this could happen. So the klib file of this lib, which should be the output of the cinterop task I guess, is also marked as its input. Input property 'libraries' file /Users/natario /Projects/DM /dm-effects/effects-backend /build/classes /kotlin/androidNativeArm64 /main/effects-backend-cinterop-gles31.klib has been added. However gradle says Task ':effects-backend:cinteropGles31AndroidNativeArm64' is not up-to-date because: Run task cinteropGles31 again Note that this task should not even run! It should be UP-TO-DATE. 71 abi_version= 22 exportForwardDeclarations=cnames.structs._GLsync 0 interop= true linkerOpts=-lGLESv3Ĭompiler_version= 1.3. klib/manifest is: unique_name=effects-backend-cinterop-gles31 Clean environment, run task cinteropGles31. Focusing on the single cinterop library called Gles31 case for simplicity.


 0 kommentar(er)
0 kommentar(er)
